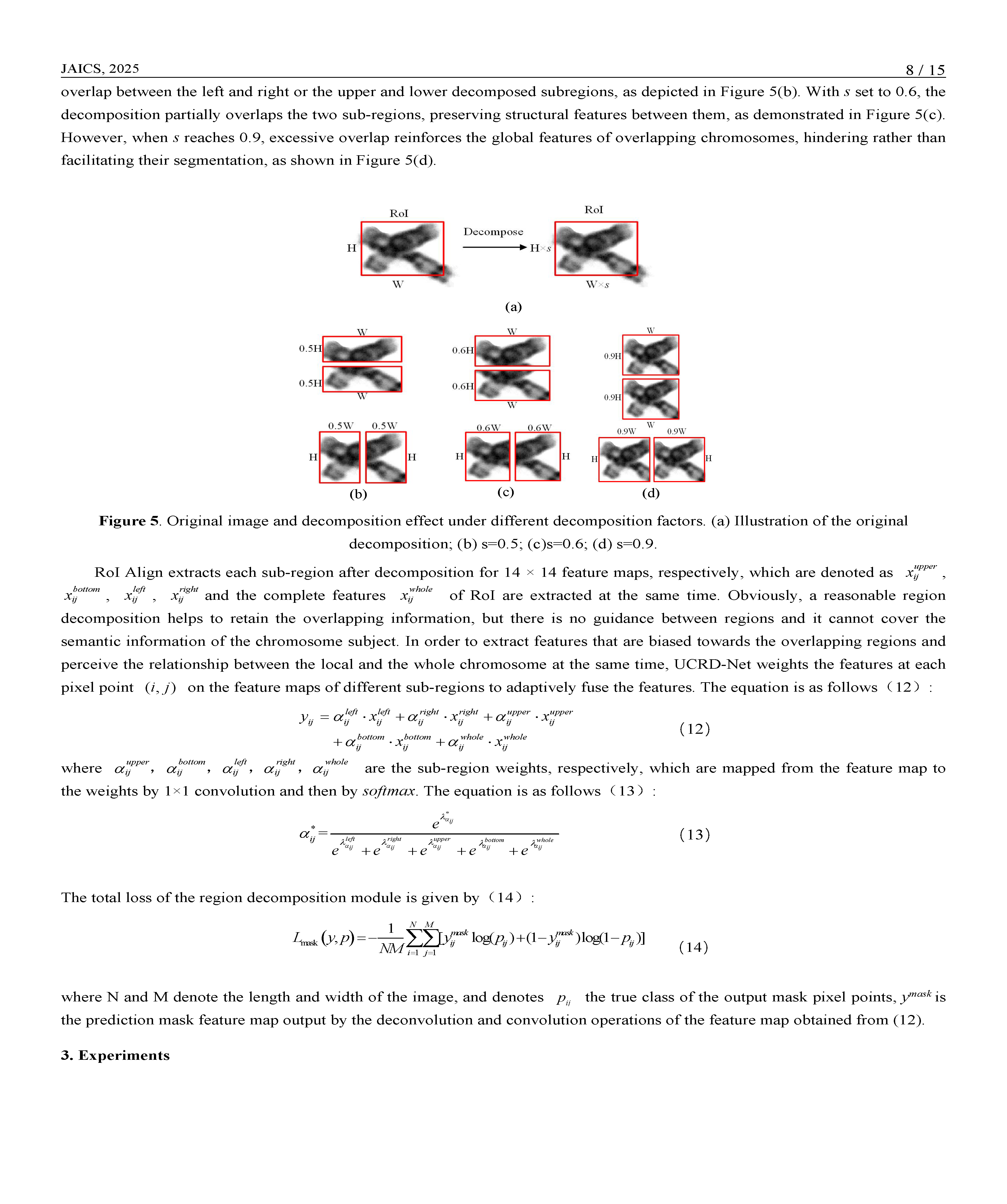
Chromosome segmentation is the basis of karyotype analysis, and the performance of segmentation directly affects the subsequent classification and identification. However, chromosomes are free in cells, presenting as non-rigid and highly susceptible to overlapping adhesions, which leads to the problems of inaccurate instance detection and incomplete segmented masks in chromosome instance segmentation. In this context, we develop an end-to-end chromosome instance segmentation network, named UCRD-Net, using uncertainty correction and region decomposition to detect and segment more accurately. In this model, the output uncertainty of the uncertainty branch is designed to correct the original classification confidence by Gaussian modeling the detection box of the chromosome instance to improve the positioning accuracy of the detection box. A strategy of decomposing Region of Interest (RoI) is proposed to weaken the semantic ambiguity of overlapping chromosomes in RoI and strengthen the characteristic response of chromosomes to be segmented. Based on the above ideas, this study has completed the end-to-end architecture design. Experiments on clinical datasets show that the average segmentation accuracy of UCRD-Net is 4.1% higher than that of the baseline model Mask R-CNN, AP50 is 0.4% higher, and AP75 is 1.0% higher. The improved accuracy and completeness of segmentation provided by UCRD-Net can offer reliable support for downstream karyotype analysis and facilitate the early detection of chromosomal abnormalities in clinical diagnostic workflows